Deciphering Gene Regulatory Networks
Every cell in your body – from liver cells to skin cells and everything in between – holds the same genetic information, the programming that tells cells what to become and how to perform the appropriate functions. So how is it that you end up with such a variety of cells capable of vastly different things?
The key is regulation of the enormous amount of genetic information contained in each cell. Different types of cells read different parts of the genetic program; in cells that will become part of the liver, the genes that code for the structure and function of liver cells are turned on – or expressed – while the genes that prescribe the features of skin cells are turned off.

Sushmita Roy
Throughout an organism’s development, cells progress from one type to another, differentiating toward specific fates. Understanding how one cell type becomes another cell type means understanding the molecular machinery responsible for every step and stage of differentiation. Unfortunately, regulation of gene expression is not as simple as an on/off switch.
Rather, vast and complex networks underlie cell lineages, dictating the nature of various cell states and the transitions between them. Sushmita Roy, Assistant Professor of Biostatistics and Medical Informatics in the Systems Biology theme at the Wisconsin Institute for Discovery, uses innovative statistical computational methods to determine what these networks are and how they might be useful for solving related biological and biomedical problems.
“The way we think about this problem is that there is a network that connects all of the genes together. It’s not one network, there are perhaps many variations to this network, that specify how gene expression happens,” says Roy. “Our goal is to try to find what these networks are, how they drive downstream gene expression, and how these networks change across different cell types, tissues, disease conditions, and across different species.”
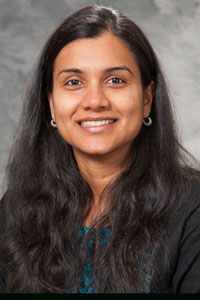
Rupa Sridharan
Learning more about gene regulatory systems is of great interest to many researchers, especially those who are trying to manipulate cells toward particular states. One such researcher is Rupa Sridharan, Assistant Professor of Cell and Regenerative Biology in the Epigenetics theme at WID. Sridharan’s research on reprogramming cells concentrates on converting differentiated or somatic cells into pluripotent stem cells, which then have the potential to re-differentiate into any cell type.
But exactly what on the molecular level makes a stem cell a stem cell is not completely clear. The identity of a cell type relies on which genes are expressed, meaning it is dependent on the regulatory networks Roy is studying. A network roadmap could help Sridharan to better understand and more efficiently facilitate cell reprogramming. With such a roadmap, according to Sridharan, “we would have the blueprint of what makes a stem cell a stem cell,” a potentially significant tool for use in regenerative medicine.
A recently awarded grant from the National Institutes of Health (NIH) brings Roy’s computational expertise to bear on the biological problems facing researchers like Sridharan who want to learn something about particular cell types and populations, while in turn providing Roy with the experimental data she needs to better characterize regulatory networks in mammalian systems, namely in mouse and human cell types.
“We can measure the components of the circuitry, but figuring out the connections among the different molecules is a hard problem – we are trying to reverse engineer what the network might be.”
-Sushmita Roy
It takes a lot of data to create useful models, but through a number of collaborative projects with Sridharan, Morgridge Institute for Research Regenerative Biology Director James Thomson, and others, along with publicly available resources, Roy has assembled rich datasets containing measurements of the numerous important network components: transcription factors, signaling proteins, three-dimensional genome organization, and many others.
With these data sets in hand, Roy can deploy her statistical and computational methods to reconstruct gene regulatory networks. “We can measure the components of the circuitry, but figuring out the connections among the different molecules is a hard problem – we are trying to reverse engineer what the network might be based on these different measurements,” says Roy.
To tackle that problem, Roy must consider each step of the changes cells go through as they develop. The molecular measurements in Roy’s amassed datasets include measurements taken at multiple time points as cells gradually differentiate and turn into different cell types. As the relationships between molecular components change, the interplay between them begins to emerge as a cohesive network.
Owing to the amount of data available, commonly used and studied cell types’ regulatory networks and how they establish global gene expression can be fairly comprehensively characterized. Those networks can be put to use in constructing other networks.
Working with biologists like Sridharan, Roy may be able to learn a great deal about the regulatory networks of novel and rare cell types by looking for overlap with existing networks. She can also use the previously collected data to make predictions about new networks, relying on her collaborators’ experimental expertise.
Newly-constructed model regulatory networks can be used as hypothesis generators, helping to determine what kinds of experiments will be most useful for answering regenerative biologists’ most pressing questions. “Once the networks have been built for different cell types and we can figure out what changes at the network level between different cell types,” says Roy, “these changes are potential predictions of what is driving a particular cell type or cell population and then we can use that to prioritize predictions.”
By examining the effects of environmental changes and perturbations on a given network, Roy and her collaborators will be able to determine what molecular measurements are most important for learning more about a cell state or population, making the lab experiments performed by Sridharan and other biologists much more targeted and efficient.
“I hope that this will be a seed to do something bigger between groups who are interested in understanding how regulatory networks establish cell fate.”
-Sushmita Roy
The project is in its early months, but in many ways it is a continuation of existing collaborations that span several UW-Madison departments and multiple WID research themes. Roy and Sridharan, for example, have been discussing gene regulatory networks ever since they both joined WID in 2012. “This has been a very good collaboration because we are very complementary: I can’t do what [Sushmita Roy] does and she can’t do what I do… and she is very willing to listen to the biologist’s perspective – she is very motivated for [her work] to be useful to biologists,” making the oft difficult task of working across disciplines seem perfectly natural.
Roy is hopeful that this project is only the beginning of this kind of collaborative effort: “I hope that this will be a seed to do something bigger between groups who are interested in understanding how regulatory networks establish cell fate,” says Roy. The new NIH project will not provide all the answers about the complex molecular systems underlying gene expression and cell lineages, but it represents an important step forward and builds the foundations of the cross-disciplinary teams needed to discover those answers.
– Nolan Lendved