Deciphering the Regulatory Network of a Pathogenic Fungus

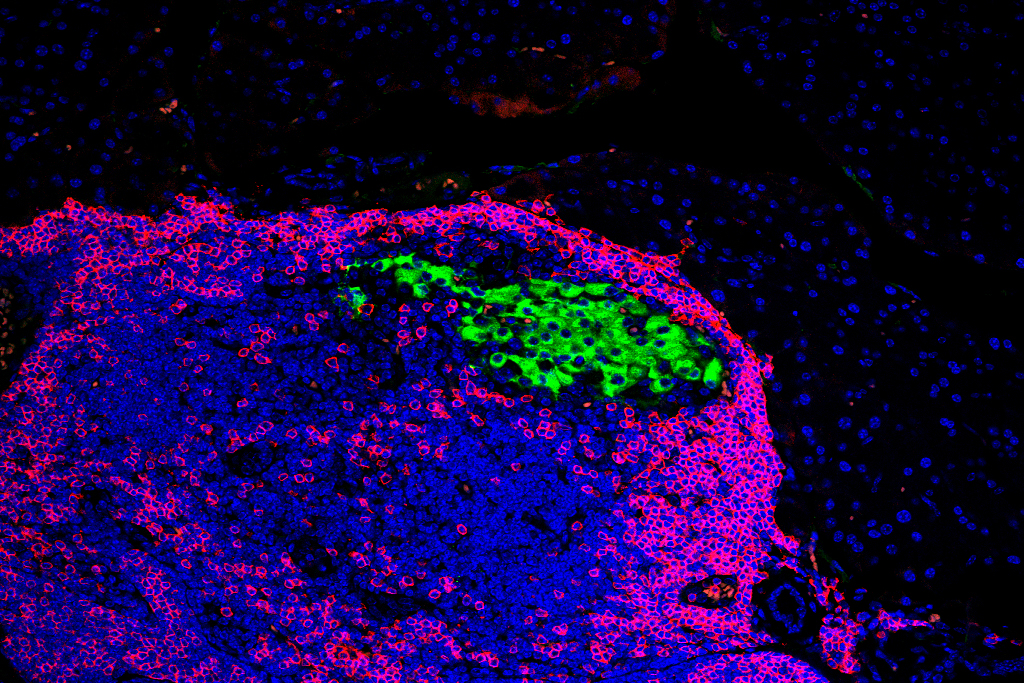
Image credit: Katherine Baldwin
In a new study, researchers from the Wisconsin Institute for Discovery (WID) (Sushmita Roy lab), in collaboration with the Department of Bacteriology and Department of Medical Microbiology and Immunology at the University of Wisconsin–Madison (Jean-Michel Ane and Nancy Keller labs) have created a software tool that can help reveal biological pathways of a notorious pathogenic fungus. Aspergillus fumigatus, which is found worldwide, can infiltrate a human body and quickly overwhelm the immune system. In immunocompromised individuals, this fungus can cause major damage and has a high mortality rate. The new tool may eventually help researchers address the problems caused by A. fumigatus.
A. fumigatus is responsible for various harmful, sometimes lethal, diseases known as aspergilloses. Most people breathe in A. fumigatus spores, commonly found in soil, decaying leaves, compost, dust, and indoor air, every day without getting sick. However, when the immune system is weakened by asthma, chemotherapy, or lung damage from an underlying disease, A. fumigatus can cause serious illness, including invasive aspergillosis that is treated by antifungal medications. Treatments include antifungal medications and, in severe cases, surgery to remove fungal balls from lung tissue.
In October 2022, the World Health Organization released its first-ever list of priority fungal pathogens, highlighting fungi that pose the greatest threat to human health. A. fumigatus was placed in the highest-risk category, alongside Cryptococcus neoformans, Candida albicans, and Candida auris. This designation reflects a sharp rise in severe aspergillosis cases and deaths, increased co-infections with COVID-19 and other respiratory illnesses, and growing resistance to antifungal treatments.
In order to comprehend the molecular mechanisms that govern A. fumigatus’ diverse life phases, researchers examined the gene regulatory networks to better understand how the fungus grows, develops, and becomes harmful. Utilizing 18 publicly available RNA-seq datasets of A. fumigatus, researchers built a comprehensive network resource, dubbed GRAsp (Gene Regulation of Aspergillus fumigatus), to examine different sets of regulatory relationships involved in specific biological processes. Drawing on the wealth of existing datasets, they constructed a genome-scale gene regulatory network (GRN) that is easily accessible to the research community.
GRAsp visualizes and explores predicted GRNs and facilitates model-guided hypothesis generation and experimental designs. It studies the underlying phenotypes of the fungus and helps explain processes involved in growth, development, and the traits that turn it into a pathogen.
GRAsp was developed as a user-friendly web portal that allows users to query the inferred GRN with individual genes and gene sets. The tool identifies new genes involved in different biological pathways by grouping related genes, finding key connections between them, and following the spread of activity across the network. Researchers then tested GRAsp in two real-world cases: they successfully uncovered a new regulator that keeps a harmful fungal toxin (gliotoxin) in check, and demonstrated that a protein called AtfA plays a key role in how fungi respond to chemical signals from other microbes and adjust how they grow or act in response.

Sushmita Roy, WID Faculty
While this work represents a significant advance, the researchers see clear opportunities to strengthen the model as new data become available and our understanding of this fungus continues to grow. “Collecting new gene expression and other omic datasets beyond gene expression, that are informed by our initial model predictions, could significantly improve our model’s ability to make in silico predictions of previously unseen perturbations,” says Sushmita Roy, WID faculty and professor of biostatistics and medical informatics. As new expression datasets become publicly available, they can be incorporated into the framework to refine existing pathways and potentially uncover new ones. The inferred GRN and associated GRAsp tool are expected to be valuable resources for identifying key genes and pathways underlying the pathogenic traits of A. fumigatus.
By making GRAsp publicly available, the team equips researchers with a powerful new way to study how this fungus functions and to pinpoint the genes and pathways that enable it to grow, survive stress, and cause disease. “Better tools that can predict the impact of previously unseen perturbations by building such network models that are also interpretable could help identify potential vulnerable pathways and help prepare for better therapeutic strategies,” says Roy.
-Laura RedEagle