In the earliest stages of life, just as cells are dividing and setting out on their paths to become liver cells, skin cells, neurons, and every other kind of cell, an intricate gene regulatory network rules development. While every cell has the same DNA, their fate depends on which portions are expressed, or turned on. The network defines interactions between genes and regulatory proteins called transcription factors—the chemical knobs that control whether and how genes are expressed— and guides early embryonic neurodevelopment.
This month, the labs of Associate Professor of Biomedical Engineering Randolph Ashton and Associate Professor of Biostatistics and Medical Informatics Sushmita Roy published a paper in Cell Systems that describes a previously unknown link between a transcription factor and an early neurodevelopmental gene. The discovery was made possible through a combination of computational and wet-lab experimental techniques and provides insights to how early stem cells develop into neurons during human development by exploring regulator-gene dynamics.

Sushmita Roy 
Randolph Ashton
Both the regulator, named POU3F2, and gene, labeled STMN2, have known roles in neurodevelopment. POU3F2 is key during early motor neuron specification, helping to turn stem cells into the nerves that control movement. STMN2 has roles in neuronal growth, cellular structure dynamics, and cellular migration during early development. Its functions extend into adulthood, however, and because of its role in maintaining cellular integrity, dysregulation of STMN2 has been associated with neurodegenerative diseases, including Rett syndrome and amyotrophic lateral sclerosis (ALS).
Finding the connection between POU3F2 and STMN2 was only possible because of a connection between scientists from disparate disciplines, a specialty of the Wisconsin Institute for Discovery, where both Ashton and Roy hold appointments. “The computational people can’t be doing things alone; experimentalists cannot be doing things alone. Inferring these gene regulatory networks requires a tight collaboration between experimental and computational labs and it requires a lot of resources and commitment from both ends—people, money and time—to be able to do this,” says Roy. “Both our labs offer complementary tools to answer this bigger question of figuring out what the regulatory network is,” she adds.
Inferring these gene regulatory networks requires a tight collaboration between experimental and computational labs and it requires a lot of resources and commitment from both ends.
— Sushmita Roy
And the question is indeed big. Scientists have long been interested in studying early human neurodevelopment, but access to embryonic human tissues is both practically and ethically challenging. However, over the past few years, researchers in the Ashton and Roy labs have created novel cellular models of early human neurodevelopment as well as computational tools to associate transcription factors with the genes they regulate. The collaborative project is funded by the Environmental Protection Agency and the National Institutes of Health.
The human nervous system begins with a region-specific cylindrical tube of neural stem cells (NSCs) that become the brain and spinal cord tissue. Growth of this tube is guided by various proteins—or growth factors—present at different levels across time. The Ashton lab has developed a protocol for cultivating human stem cells—capable of developing into any cell type—into NSCs by exposing them to different growth factors at different times. A strong collaboration with Professor James Thomson, Director of Regenerative Biology at the Morgridge Institute for Research, has been critical for taking the next steps: capturing gene expression data over time as the cells differentiate and begin to resemble those present in the posterior human neural tube.
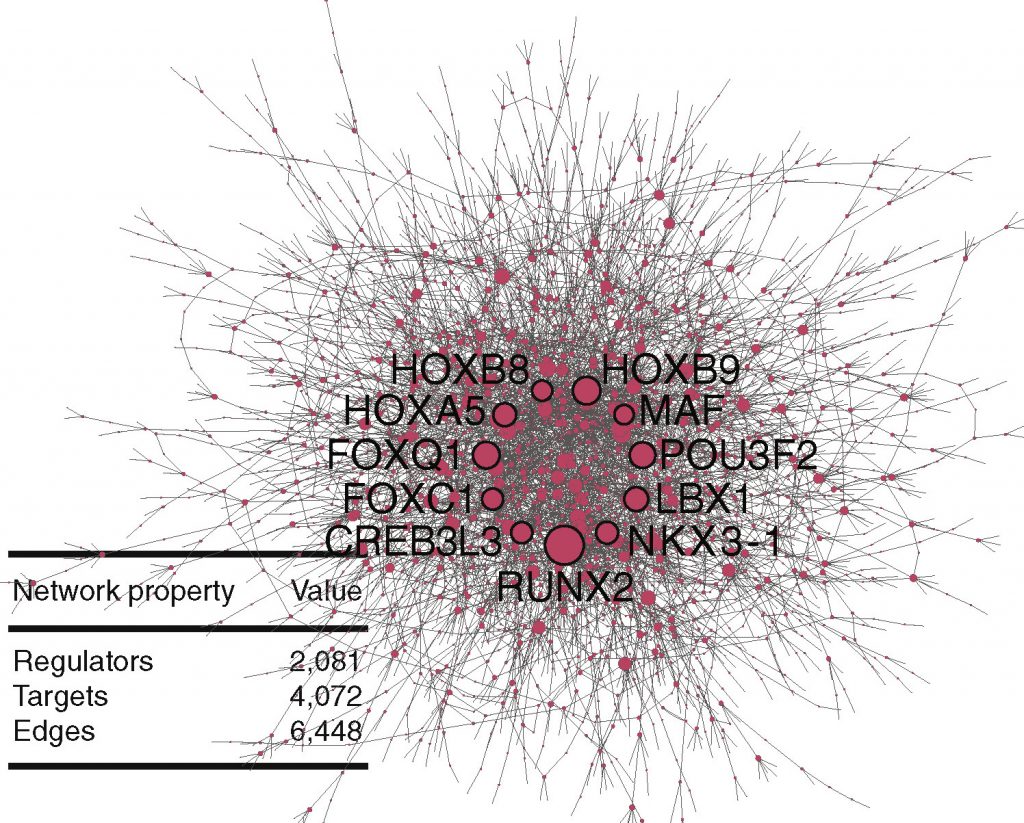
With the time series data in hand, Deborah Chasman, a postdoctoral research associate formerly working in the Roy lab, developed an algorithm to predict dynamic genes—genes in which expression levels change over time and which are responsible for cellular differentiation. But the data from the Ashton and Thomson labs’ experiments was not enough to bring the network into focus. Thankfully, gene expression data sets generated as part of any published experimental study is publicly available through databases such as Gene Expression Omnibus (GEO) or ArrayExpress. Another computational tool developed by graduate student Alireza Fotuhi Siahpirani in the Roy lab, called MERLIN-P, combined related publicly-available data with the experimental data from the Ashton and Thomson labs to uncover possible connections between transcription factors and their target genes. The result was a newly-described gene regulatory network.

Deborah Chasman 
Nisha Iyer
MERLIN-P identified several regulators of the network. Ashton lab postdoctoral researcher Nisha Iyer investigated the supposed transcription factor and gene pair findings with a series of experiments that tested whether the identified regulators acted upon their network target gene. That’s when the group discovered the novel regulatory relationship between POU3F2 and STMN2. “[Finding this connection] was cool because both of these genes have been shown to be involved in the early brain development, but nobody’s actually connected them together—we were able to establish this regulatory connection,” Roy says.
Roy notes that their findings only scratch the surface of understanding gene regulatory networks. But with continued collaboration, more of these discoveries are possible. “There are thousands of [connections] and we’ve only validated four or five of them. It’s a very small step, but I think this is how we should be doing things,” she says.
This work was made possible in part by USEPA grant 83573701 to Sushmita Roy and Randolph Ashton, NIH NIGMS grant 1R01GM117339 to Sushmita Roy, and a grant from Marv and Mildred Conney to Randolph Ashton and James Thomson.
You must be logged in to post a comment.