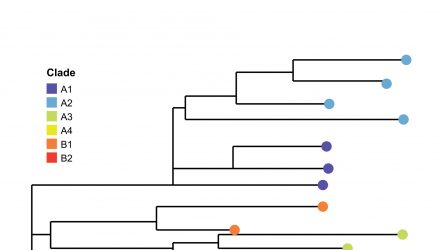
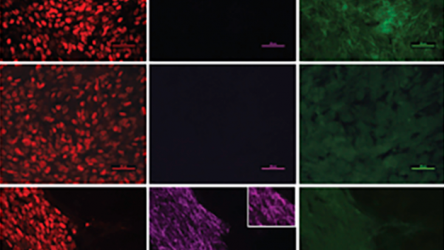
Ashton Group: Stem Cell Bioprocessing and Regenerative Biomaterials Lab
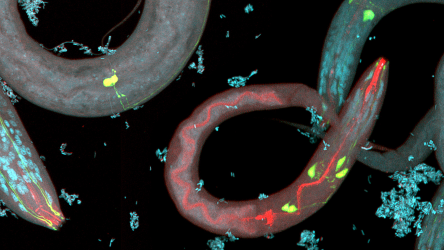
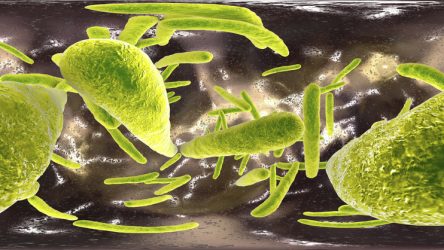
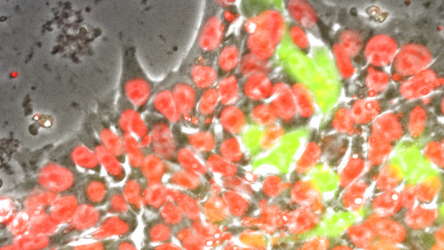
The Ashton Group is working to understand, model, and recapitulate in vitro the instructive signals utilized by human embryos to pattern tissue-specific differentiation of pluripotent stem cells, and apply this knowledge towards the rational design of tissue engineered scaffolds and other regenerative therapeutic strategies.