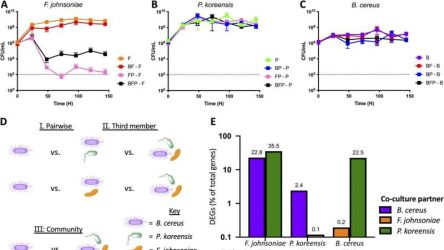
We Are About to Enter the Golden Age of Gene Therapy
From the Inverse: Krishanu Saha, a bioengineer at the University of Wisconsin–Madison whose lab is working on gene therapies for treating blindness, says the precision allowed by CRISPR-Cas9’s programmability is its singular selling point.