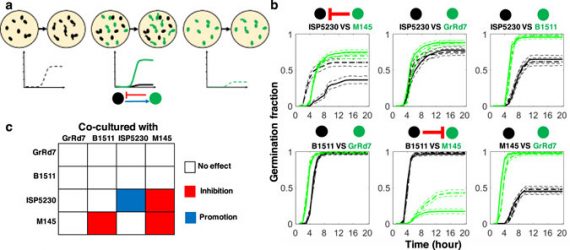
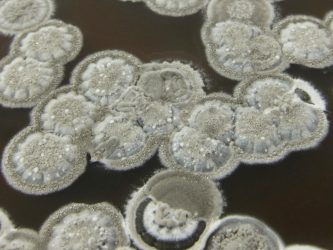
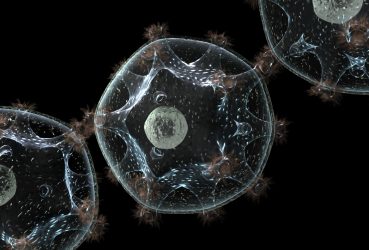
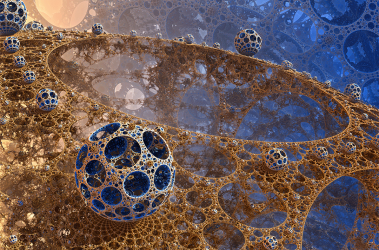
Phenotypic variability and community interactions of germinating Streptomyces spores
Systems Biology researcher Kalin Vetsigian and graduate student Ye Xu recently published findings in Nature’s Scientific Reports about the stochasticity of growth within Streptomycetes spore communities.